Contents
U-Net LAB
Learn how to to do U-Net
================= Load Keras Modules=================
#Import Keras Modules
import os
import random
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
plt.style.use("ggplot")
%matplotlib inline from tqdm import tqdm_notebook, tnrange
from itertools import chain
from skimage.io import imread, imshow, concatenate_images
from skimage.transform import resize
from skimage.morphology import label
from sklearn.model_selection import train_test_split
from keras.models import Model, load_model
from keras.layers import Input, BatchNormalization, Activation, Dense, Dropout
from keras.layers.core import Lambda, RepeatVector, Reshape
from keras.layers.convolutional import Conv2D, Conv2DTranspose
from keras.layers.pooling import MaxPooling2D, GlobalMaxPool2D
from keras.layers.merge import concatenate, add
from keras.callbacks import EarlyStopping, ModelCheckpoint, ReduceLROnPlateau
from keras.optimizers import Adam
from keras.preprocessing.image import ImageDataGenerator, array_to_img, img_to_array, load_img
from keras import initializers
================= Load Images and Labels=================
# Set input width and height
im_width = 128
im_height = 128
# lload all the images in images folder # The name of images are stored in ids ids = next(os.walk("images"))[2]
print("No. of images = ", len(ids)) # Allocate the space for training and label X = np.zeros((len(ids), im_height, im_width, 1), dtype=np.float32)
y = np.zeros((len(ids), im_height, im_width, 1), dtype=np.float32) # Load the all the images and labels, the labels are called as masks here
for n in range(len(ids)):
# Load images # img_to_array: convert image to array
img = load_img("images/"+ids[n], grayscale=True)
x_img = img_to_array(img)
x_img = resize(x_img, (128, 128, 1), mode = 'constant', preserve_range = True)
# Load masks
mask = img_to_array(load_img("masks/"+ids[n], grayscale=True))
mask = resize(mask, (128, 128, 1), mode = 'constant', preserve_range = True)
# Normalization image
X[n] = x_img/255.0
y[n] = mask/255.0 # Split all the images into training data and validation data # test_size 0.2 means 20% valication set 80% training set # random state: random seed
X_train, X_valid, y_train, y_valid = train_test_split(X, y, test_size=0.2, random_state=42)
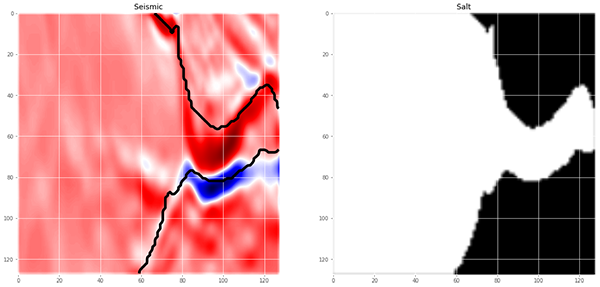
================= Visualize Images and Labels=================
# Visualize any random image and the mask in the training data
ix = random.randint(0, len(X_train)) # Plot seismic image
fig, (ax1, ax2) = plt.subplots(1, 2, figsize = (20, 15))
ax1.imshow(X_train[ix, ..., 0], cmap = 'seismic', interpolation = 'bilinear')
# If there is salt in the image, plot the boundary of salt as black line
has_mask = y_train[ix].max() > 0 # salt indicator
if has_mask: # if salt
# draw a boundary(contour) in the original image separating salt and non-salt areas
ax1.contour(y_train[ix].squeeze(), colors = 'k', linewidths = 5, levels = [0.5])
ax1.set_title('Seismic') # Plot salt labels ax2.imshow(y_train[ix].squeeze(), cmap = 'gray', interpolation = 'bilinear')
ax2.set_title('Salt')
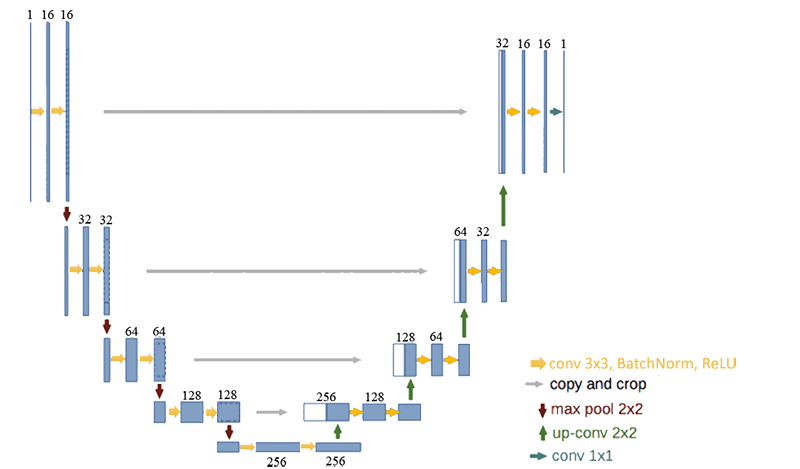
================ Setup U-Net Structure================
#define the convolution layer def conv2d_block(input_tensor, n_filters, kernel_size = 3, batchnorm = True):
"""Function to add 2 convolutional layers with the parameters passed to it"""
# first convolution layer
x = Conv2D(filters = n_filters, kernel_size = (kernel_size, kernel_size),\
kernel_initializer = initializers.he_normal(seed=1), padding = 'same')(input_tensor)
if batchnorm:
x = BatchNormalization()(x)
x = Activation('relu')(x)
# second convolution layer
x = Conv2D(filters = n_filters, kernel_size = (kernel_size, kernel_size),\
kernel_initializer = initializers.he_normal(seed=1), padding = 'same')(x)
if batchnorm:
x = BatchNormalization()(x)
x = Activation('relu')(x)
return x # define U_net architecture def get_unet(input_img, n_filters = 16, dropout = 0.1, batchnorm = True):
"""Function to define the UNET Model"""
# Contracting Path
c1 = conv2d_block(input_img, n_filters * 1, kernel_size = 3, batchnorm = batchnorm)
p1 = MaxPooling2D((2, 2))(c1)
p1 = Dropout(dropout)(p1)
c2 = conv2d_block(p1, n_filters * 2, kernel_size = 3, batchnorm = batchnorm)
p2 = MaxPooling2D((2, 2))(c2)
p2 = Dropout(dropout)(p2)
c3 = conv2d_block(p2, n_filters * 4, kernel_size = 3, batchnorm = batchnorm)
p3 = MaxPooling2D((2, 2))(c3)
p3 = Dropout(dropout)(p3)
c4 = conv2d_block(p3, n_filters * 8, kernel_size = 3, batchnorm = batchnorm)
p4 = MaxPooling2D((2, 2))(c4)
p4 = Dropout(dropout)(p4)
c5 = conv2d_block(p4, n_filters = n_filters * 16, kernel_size = 3, batchnorm = batchnorm)
# Expansive Path
u6 = Conv2DTranspose(n_filters * 8, (3, 3), strides = (2, 2), padding = 'same')(c5)
u6 = concatenate([u6, c4])
u6 = Dropout(dropout)(u6)
c6 = conv2d_block(u6, n_filters * 8, kernel_size = 3, batchnorm = batchnorm)
u7 = Conv2DTranspose(n_filters * 4, (3, 3), strides = (2, 2), padding = 'same')(c6)
u7 = concatenate([u7, c3])
u7 = Dropout(dropout)(u7)
c7 = conv2d_block(u7, n_filters * 4, kernel_size = 3, batchnorm = batchnorm)
u8 = Conv2DTranspose(n_filters * 2, (3, 3), strides = (2, 2), padding = 'same')(c7)
u8 = concatenate([u8, c2])
u8 = Dropout(dropout)(u8)
c8 = conv2d_block(u8, n_filters * 2, kernel_size = 3, batchnorm = batchnorm)
u9 = Conv2DTranspose(n_filters * 1, (3, 3), strides = (2, 2), padding = 'same')(c8)
u9 = concatenate([u9, c1])
u9 = Dropout(dropout)(u9)
c9 = conv2d_block(u9, n_filters * 1, kernel_size = 3, batchnorm = batchnorm)
outputs = Conv2D(1, (1, 1), activation='sigmoid')(c9)
model = Model(inputs=[input_img], outputs=[outputs])
return model
=================== Training U-Net ===================
# Set input layer size input_img = Input((im_height, im_width, 1), name='img') # Set the parameters for Unet
model = get_unet(input_img, n_filters=16, dropout=0.05, batchnorm=True) # Set optimizer, loss function. Set accuracy as metric
model.compile(optimizer=Adam(), loss="binary_crossentropy", metrics=["accuracy"]) #Visualize the structure of Unet model.summary() # Set criterias for the inversion procedure
# Early Stopping: If the validation set loss function does not decrease in 10 iteration, then early stop.
# ReduceLROnPlateau: If the validation set loss function does not decrease in 5 iteration, then multiply learning rate with factor. # ModelCheckpoint: save_weights_only=True--Save only the weight,
#save_best_only=True -- Only Save the best model with lowest validucation loss
#'model-tgs-salt.h5' -- file name callbacks = [
EarlyStopping(monitor='val_loss',patience=10, verbose=1),
ReduceLROnPlateau(factor=0.1, patience=5, min_lr=0.00001, verbose=1), ModelCheckpoint('model-tgs-salt.h5', verbose=1, save_best_only=True, save_weights_only=True)
] # Train the network
# Set the input
# batch_size--Size for each mini batch
# epochs--Epoch number
# X_train, y_train--Training data and labels
# X_valid, y_valid--Validation data and labels results = model.fit(X_train, y_train, batch_size=32, epochs=30, callbacks=callbacks,\ validation_data=(X_valid, y_valid))
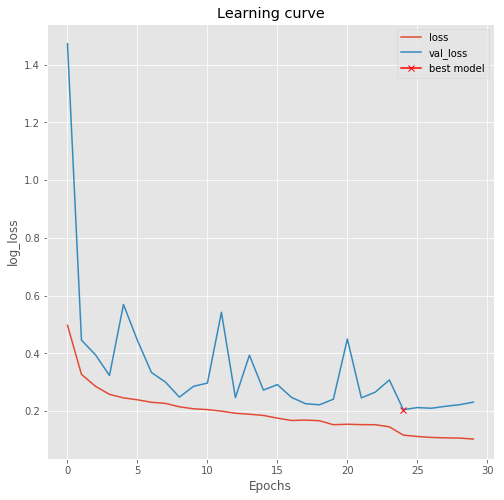
================= Plot Learning Curve=================
# plot learning curve plt.figure(figsize=(8, 8))
plt.title("Learning curve")
plt.plot(results.history["loss"], label="loss")
plt.plot(results.history["val_loss"], label="val_loss") # plot the location of the best model
plt.plot( np.argmin(results.history["val_loss"]), np.min(results.history["val_loss"]), marker="x", color="r", label="best model")
plt.xlabel("Epochs")
plt.ylabel("log_loss")
plt.legend();
================= Implement Model on Validation Set =================
# load the best model
model.load_weights('model-tgs-salt.h5')# Evaluate on validation set loss and accuracy
model.evaluate(X_valid, y_valid, verbose=1)# Predict on training and validation set
preds_train = model.predict(X_train, verbose=1)
preds_val = model.predict(X_valid, verbose=1)# Threshold predictions
preds_train_t = (preds_train > 0.5).astype(np.uint8)
preds_val_t = (preds_val > 0.5).astype(np.uint8)
================= Implement Model on Validation Set =================
# Plot predict resultsdef plot_sample(X, y, preds, binary_preds, ix=None):
"""Function to plot the results""
# ix is None, then random select image to plot
if ix is None:
ix = random.randint(0, len(X))fig, ax = plt.subplots(1, 4, figsize=(20, 10))
# plot input seismic image
ax[0].imshow(X[ix, ..., 0], cmap='seismic')# If there is salt in the image, plot the boundary of salt as black line
has_mask = y[ix].max() > 0
if has_mask:
ax[0].contour(y[ix].squeeze(), colors='k', levels=[0.5])
ax[0].set_title('Seismic')# plot true salt labels
ax[1].imshow(y[ix].squeeze())
ax[1].set_title('Salt')# Plot predicted salt probability ax[2].imshow(preds[ix].squeeze(), vmin=0, vmax=1)
if has_mask:
ax[2].contour(y[ix].squeeze(), colors='k', levels=[0.5])
ax[2].set_title('Salt Predicted')# Plot predicted salt Labels
ax[3].imshow(binary_preds[ix].squeeze(), vmin=0, vmax=1)
if has_mask:
ax[3].contour(y[ix].squeeze(), colors='k', levels=[0.5])
ax[3].set_title('Salt Predicted binary');# Check if training data looks all right
plot_sample(X_train, y_train, preds_train, preds_train_t, ix=14)plot_sample(X_train, y_train, preds_train, preds_train_t)
# Check if validation data looks all right
plot_sample(X_valid, y_valid, preds_val, preds_val_t)
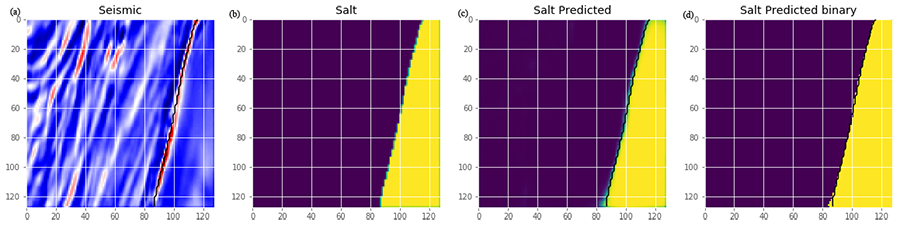
Figure 4: (a) Input seismic image, (b) true salt labels, (c) perdicted salt probabilities and (d) predicted salt labels
The code above adapter from https://github.com/hlamba28/UNET-TGS